You are here
Fungal Effectors
H. vastatrix transcriptomic studies were conducted using 454-pyrosequencing and RT-qPCR approaches allowing the characterization of 9234 putative fungal genes revealing :
- Intense signalling, transport and secretory activity in germ tubes suggesting the onset of a plant-fungus dialogue at this early stage of infection;
- Up-regulation of biogenesis, lipid transport and energy production during apressorium formation;
- Increase of DNA replication and cell division transcripts at the onset of sporulation;
- 516 genes were predicted to encode secreted proteins (candidate effectors), particularly in germ tubes and haustoria.
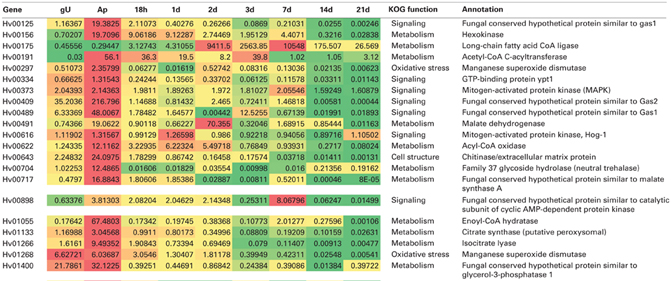
Talhinhas et al. (2014) Frontiers in Plant Science 5:88: (doi: 10.3389/fpls.2014.00088)
As a preliminary step towards sequencing the genome of H. vastatrix (on-going work), the genome size estimated at 797 Mbp was measured by flow cytometry. This approach was subsequently applied to a range of other rust fungi, showing that rusts (Pucciniales) are, as a group, the organisms with the largest average genome size in the kingdom Fungi.
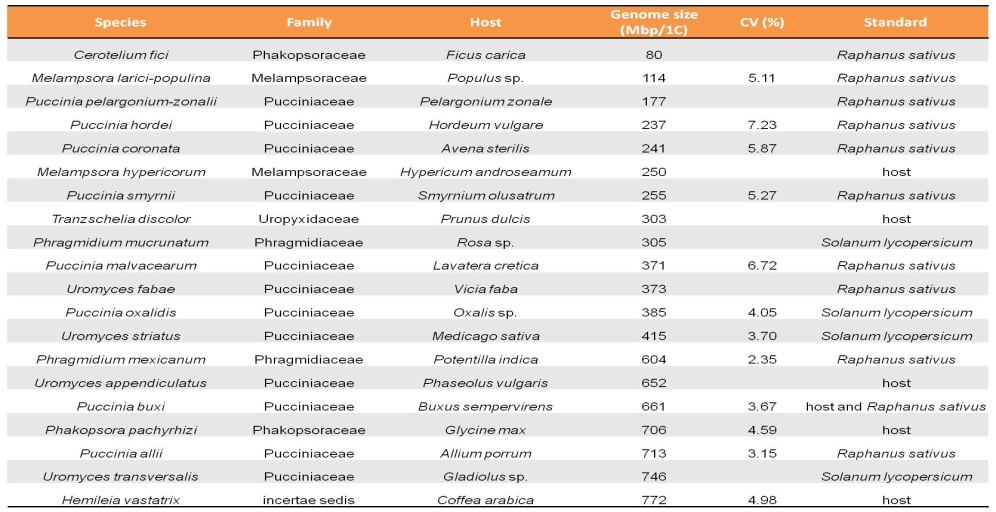
Tavares et al. (2014). Front. Plant. Sci 5:422 (doi: 10.3389/fpls.2014.00422)
A combination of karyological and genomic tools were used to characterize differences among C. kahawae isolates concerning the genome size, which ranged 74-89 Mb, and the number of minichromosomes (below 2 Mb), from 1 to 5. A positive correlation could be established between the number of minichromosomes and the aggressiveness.
Selected references:
- Talhinhas et al. (2017). Molecular Plant Pathology 18 (8): 1039–1051 (DOI: 10.1111/mpp.12512)
- Vieira et al. (2016). PLoS ONE 11 (3): e0150651. doi:10.1371/journal.pone.0150651
- Pires et al. (2016). Plant Pathol. 65: 968–977 (doi: 10.1111/ppa.12479)
- Loureiro et al. (2015). Fungal Biol.119: 1093-1099 (doi: 10.1016/j.funbio.2015.08.008)
- Tavares et al. (2014). Front. Plant. Sci 5:422 (doi: 10.3389/fpls.2014.00422)
- Talhinhas et al. (2014). Front. Plant. Sci. 5:88 (doi: 10.3389/fpls.2014.00088)
- Fernandez et al. (2012). Mol. Plant. Pathol 13: 17-37. (doi: 10.1111/J.1364-3703.2011.00723.X)



